Residual protein analysis of complex products
Regulatory authorities demand investigation, monitoring, and documentation of process-related protein residuals generated during biologics manufacturing. Such impurities can reduce drug efficacy and patient safety, and insufficient documentation may delay product approval.
Whereas traditional biologics have manufacturing processes that are well-defined and supported by a panel of established analytical test methods, the manufacturing processes for complex products, like mRNA vaccines, bacteriophages, and advanced therapies, are still in development. Therefore, improved analytical tools, such as mass spectrometry (LC-MS), are necessary to ensure process consistency and quality and to meet evolving requirements for product release and characterization.
Why use LC-MS for residual protein analysis of complex biologics?
Document process-related residual proteins
Identify and quantify cell substrate-derived, cell culture-derived, and downstream process-derived impurities
Evaluate clearance throughout purification steps
Show that your process efficiently clears process-derived impurities, resulting in a pure and safe product
Perform comparability studies
Assess protein impurity levels after process changes, upscaling, or tech transfer between CMOs to document a consistent, comparable product
Unlike traditional biologics, which are typically manufactured by expressing one recombinant protein in a single cell line, advanced therapies, viral vaccines, and oligonucleotide-based medicines contain highly heterogeneous protein mixtures from multiple sources and organisms.
Commercial HCP ELISA kits, typically used for monitoring Host Cell Proteins (HCPs) from CHO and E.coli expression systems, cannot simultaneously detect proteins from different species and often demonstrate low coverage of, e.g., HCPs from non-immunogenic human cell lines. As such, classical impurity assays often fail to detect a substantial proportion of the residual proteins in complex biotherapeutics.
Liquid chromatography-mass spectrometry (LC-MS) is a robust and reproducible orthogonal method to ELISA, which has high specificity, sensitivity, and dynamic range to detect highly heterogeneous HCPs. Using Data Independent Acquisition (DIA), LC-MS can quantify and characterize all protein impurities in a sample and provide thorough, precise, and robust HCP coverage.
Regulatory authorities often fast-track novel therapies and vaccines that have the potential to treat severe, life-threatening conditions. Furthermore, since they can be one-shot cures, being first to market can provide a competitive advantage. However, developing a process-specific ELISA for complex products requires 1 - 2 years for antibody development and assay validation. Even then, there is a risk of delayed regulatory approval due to inadequate impurity detection and clearance.
LC-MS assays require only 6-8 weeks to be established and optimized for any biologic product. This can significantly accelerate the development, marketing approval, and commercial biomanufacturing of complex biologics.
Manufacturing changes commonly happen for complex biologics, and comparability studies of residual proteins are frequently necessary for product development and regulatory filings. For instance, when improving the process consistency and product quality, upscaling, or transferring to GMP facilities for commercial release. However, demonstrating comparability in complex biologics is often challenging by still-evolving process knowledge, small numbers of batches, and material constraints for each batch.
The LC-MS assays at Alphalyse have been developed for high robustness and tolerability toward different sample types, complexity, and proteins from multiple sources. It is a flexible method allowing for process changes and the use of various buffers and matrices. The use of the same intact internal protein standards enables comparable results between samples, across process changes, and for comparability between GMP batches.
Curious to know more?

Whatever challenge or question you may have, we are here to help you solve it. One of our protein analysis experts will discuss the best analysis approach or method for your project by email or online meeting – without obligation.
Typical project process
You typically work with
these experts:



Project scope
We will start with an online meeting to learn more about your project. Based on your needs, you will receive a draft proposal outlining the suggested analyses and expected timeframe.
Samples
After signing the final project proposal, we will contact you for details about your samples. An estimated report delivery date will be sent as soon as we receive your samples.
Execution
A project leader will oversee the project and send you status updates by email at regular intervals.
The analysis varies according to the project but typically includes:
- Setting up a sequence database containing all the proteins used in your manufacturing process that could potentially end up in the drug substance.
- An optimized sample preparation developed explicitly for complex products, e.g., viral vaccines and viral vector therapies.
- Analysis using a robust microflow HPLC and SWATH® MS/MS system to ensure high reproducibility.
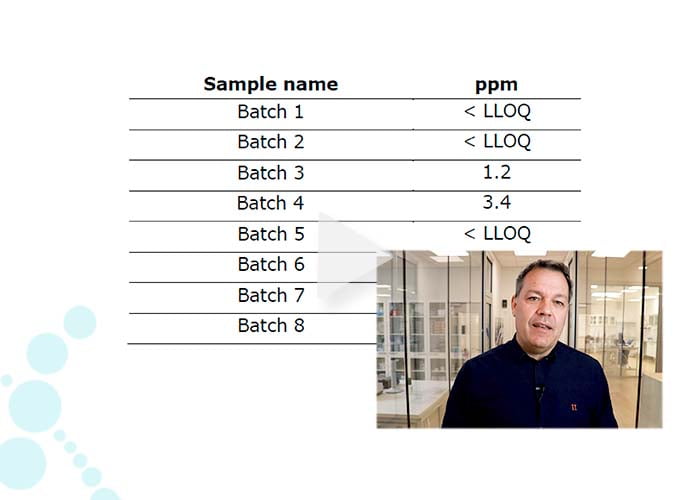
- Spike-in of internal standard proteins (0-2000 ppm), enabling quantification linearity even to low ppm levels.
Results
You will receive the analysis report by email. Depending on the project, it includes:
- Objectives, description of analytical procedure, results, and conclusions.
- List of the total HCPs in the sample.
- List of individual HCPs, process-related residuals, and their quantities.
- Selected raw data, e.g., excel sheets. You may request additional raw data if needed.
Follow up
Upon completion of the project, your team is invited to go over the results at an online meeting.
Client stories
World’s first GMP-validated MS-based residual enzyme analysis
Webinar: ELISA reagent characterization using LC-MS
Webinar: LC-MS HCP assay validation and GMP release testing
Video: ICH Q8B characterization of complex products
Webinar: Analyzing vaccine purity – without an HCP ELISA
Video: Analysis of bacteriophage product
Video: Residual biocatalysis in small molecule API
Video: Analysis of residual protein in viral products
Infographic: Analytical techniques for ensuring consistent C>s
Analysis of individual human residual proteins
Quantifying lentivirus and residual proteins in one assay
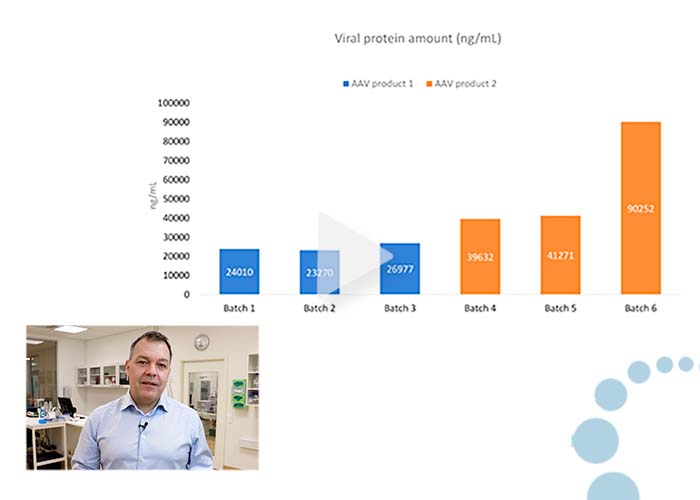
Comparing viral protein quantities in AAV batches and DS
Webinar: Monitoring impurities and ensure consistent therapies
Video: Expression systems without a commercial ELISA kit
Purified mAb with difficult-to-remove HCP impurity (ubiquitin)
Benefits of LC-MS-based impurity analysis of complex biologics
Analyze protein-based residuals from multiple organisms in one assay
Highly robust method not affected by process changes.
Includes quantification of product-specific proteins, viral proteins, etc.
Fast development of assay - within weeks.
Low amount of material required for assay development and analysis
What clients say
Knowledge center
Can residual protein testing be performed on lentiviral vectors?
Yes, and we have also performed impurity testing on other viral vectors. Viral vectors are typically highly complex and can contain proteins from multiple sources, such as the growth medium, added cellular growth factors, vector proteins, and Host Cell Proteins (HCPs) produced as part of the natural cellular metabolism.
Since viral vectors are highly complex, impurity analysis using ELISA would require several custom-made assays targeting different protein groups. Using LC-MS, it is possible to analyze both the viral vector itself and any additive material quickly, robustly, and accurately - regardless of their source.
Is it possible to analyze harvest and clarification steps?
We routinely run all types of in-process samples, from harvest to drug product and in-process samples, since our mass spectrometry-based assay works for all kinds of samples and expression systems. The sample preparation is optimized for various matrices, and only minor adjustments are needed to customize the setup for individual projects.
What are the criteria for successfully identifying a protein? Are there protein residuals that you cannot identify?
We require that at least two peptides are identified for each protein before we report the protein as identified.
The MS and MS/MS data are searched against protein sequence databases to identify peptides. We require a match for the peptide mass and peptide fragment masses for known sequences. MS-based assays achieve a very low-ppm sensitivity, making identifying even low abundant impurities possible. The method is not biased against specific proteins but requires observation of at least peptides from a protein. On request, we can set up absolute quantification MRM LC-MS for specific protein impurities of interest and perform spike-in experiments to confirm their identity.
Research article:
Videos:
- Analysis of residual protein in viral products
- Uncommon expression systems
- Analysis of bacteriophages
- ICH Q8B characterization of complex products
White paper:
Client cases:
- Comparing viral protein quantities in AAV batches and DS
- Quantifying lentivirus and residual proteins in one assay
- Orthogonal analysis of individual human residual proteins
- Analysing heterogeneous HCP mix from multiple species
- Vaccine development and process optimization
Poster:
Online articles:
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.