Literature
The mass spectrometry SWATH analysis can analyze host Cell Proteins (HCPs) in protein biopharmaceuticals. It provides both identification and quantification of the individual HCPs [1].
The analysis requires optimized sample preparation protocols for enzymatic protein cleavage into peptides. It also requires highly advanced liquid chromatography mass spectrometry (LC-MS) techniques [2].
The central analytical challenges are:
- Measurement of low-level HCPs (nanogram amount) in the presence of mg amounts of drug substance protein.
- Reproducibility of the MS techniques to identify and quantify each low-level HCP.
The solution is simple; SWATH MS analysis solves both challenges, making it an ideal tool in HCP analysis for bioprocess optimization [3].
The principles of SWATH MS analysis
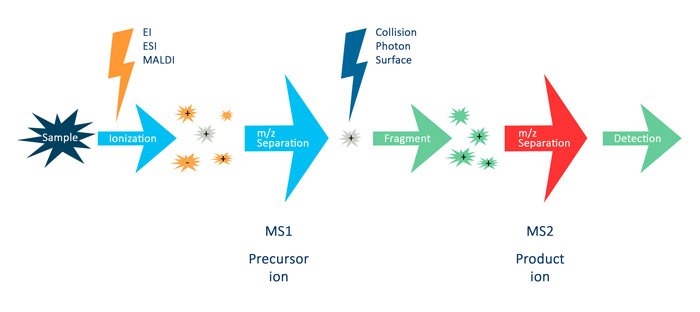
For LC-MS/MS of peptides, several terms are essential to understand:
In the figure above, the term MS (or MS1) describes the analysis where the MS instrument measures the intact mass of a peptide. MS/MS (or MS2) describes the analysis that isolates a specific peptide mass (m/z) and fragments it into specific fragment ions. You can use the mass of a peptide and its fragment ions to identify the peptide. Also, you may use the intensity of the MS signals to quantify the peptide [4, 5].
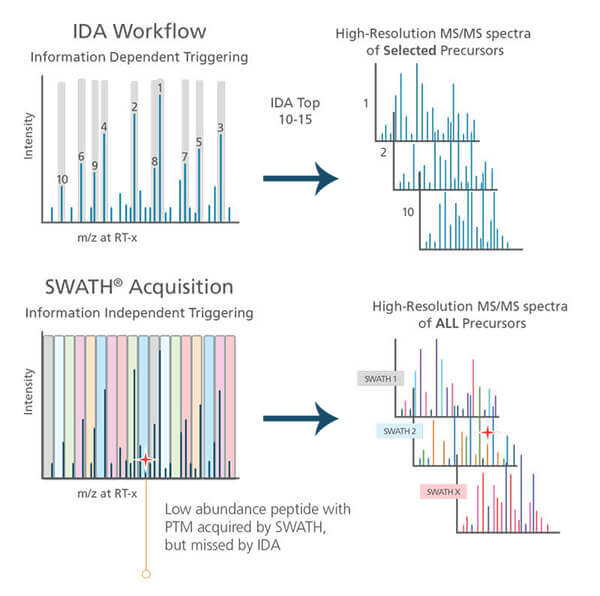
Source: sciex.com
Mass spectrometers can perform MS/MS analysis in Data Dependent Acquisition mode (DDA) [3]. DDA is sometimes called the Information Dependent Acquisition mode (IDA).
In DDA mode, the mass spectrometer first selects the most intense peptide ions (top 10-15 peaks) in MS1 and then sends them one by one into the fragmentation analysis (MS2).
Experience shows that DDA analysis of a complex peptide sample generates different protein identification results for identical technical replicates. In proteomics research, you commonly see reports of protein identification results when 2 out of 3 technical replicates observe a given protein. In the DDA mode of a complex sample, the instrument does not have time to select all peptides for MS/MS analysis. Instead, you only analyze the top 10-15 peptides. The instrument then decides which peptide to choose on the fly, creating variation from run to run [3].
Advanced mass spectrometers can also perform Data Independent Acquisition (DIA), where they fragment all peptides within a specific mass range in MS/MS. This instrument does not select individual peptides, so the data acquisition in DIA-mode is more reproducible [4, 5].
What is SWATH MS?
SWATH based mass spectrometry analysis is a particular type of DIA.
The Sciex mass spectrometer divides the mass range into small mass windows in HCP SWATH mode. Eventually, there can be up to hundreds of windows. The instrument performs MS/MS analysis on all peptides in each window. With dynamic mass windows, you will have small windows in areas with many peptides. – While larger windows in areas with few peptides [3].
“The beauty of SWATH is that you can acquire data for all peptides without knowing what to look for.”
Interpreting a complex HCP SWATH mass spectrum with the fragmentation of multiple peptides requires comparison to an ion library with mass spectrometry data on known peptides. The ion library contains information about each peptide, including HPLC retention time, MS/MS data, and amino acid sequence. The ion library is built using DDA analysis of the samples. Furthermore, you can expand it with new data on more samples [3].
Key advantages of SWATH analysis by mass spectrometry:
- Low interference from the high amount of the drug substance protein. Because you acquire the data on low-level HCP peptides in the data-independent mode in small mass windows.
- Highly reproducible HCP identification and quantification from acquiring data in Data-Independent Acquisition mode.
- All recorded SWATH MS analysis data can be re-analyzed – when updating the ion library with more known compounds and their fragmentation pattern.
Download the application note for a description of Alphalyse’s HCP analysis based on SWATH MS >>
Video: The principles of LC-MS-based HCP analysis

References
[1] Wang et al.: “Host Cell Proteins in Biologics Development: Identification, Quantitation and Risk Assessment, “Biotechnology and Bioengineering, 2009
[2] Bracewell et al.: “The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control, “Biotechnology and Bioengineering, 2015
[3] Heissel et al.: “Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein,” Protein Expression and Purification, 2018
[4] Doerr, A.: “DIA mass spectrometry,” Nature Methods, 2015
[5] Law et al.: “Recent advances in mass spectrometry: data-independent analysis and hyper reaction monitoring,” Expert Review of Proteomics,” 2013
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.
