Literature
Question:
My analytical department says mass spectrometry is impossible to run under QC conditions. Why is reproducible mass spectrometry so challenging to achieve?
Answer:
Mass spectrometry (MS)-based analysis is invaluable for many parts of the CMC documentation, including host cell protein (HCP) identification, yet reproducible quantification of proteins poses a critical challenge. The complexity and variability of MS are significant obstacles to its integration into quality control (QC) laboratories, where robustness and reproducibility are paramount.
Elements affecting the variability of MS-based host cell protein identification
Five key complications contribute to the variability challenge (figure 1):
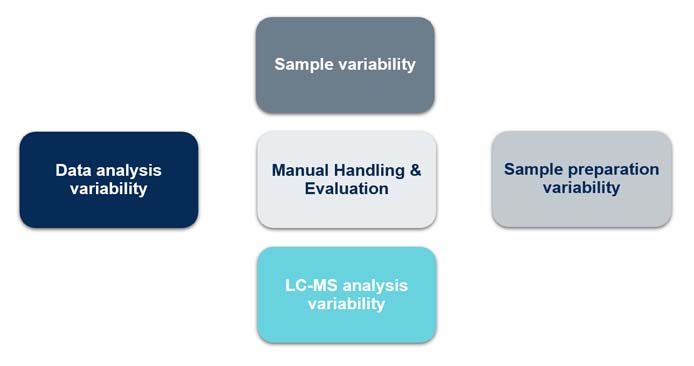
- Sample variability: Variations in protein composition, buffer composition, and concentration across samples necessitate covering a broad dynamic range (up to 6 orders of ). Most MS instruments can only cover 4 to 5 orders of magnitude.
- Sample preparation variability: Most MS-based methods involve the digestion of hundreds of proteins into thousands of peptides. Inconstant protease activity, buffer composition, potential mis-cleavages, and sample handling-induced modifications (e.g., oxidations and deamidation) all contribute to variability.
- Instrument variability: During liquid chromatographic (LC) separation, peptides elute into the MS instrument. The sensitivity of the analysis varies depending on the condition of the LC column: Whether the column is new or used can affect the chromatography results due to potential carryover from previously analyzed samples. Also, highly sensitive MS instrumentation is sensitive to contamination and will provide variable results according to the cleanliness of the instrument.
- Data analysis variability: Thousands of data points from the peptides are transformed into peak lists and searched against protein databases using diverse software, aiming to identify and quantify the peptides. Successfully translating the analytical findings to determine the identity and quantity of the original proteins in the sample depends on the quality and size of the databases and the strength and sophistication of the digital algorithms used.
- Manual handling and evaluation: All four steps above can involve more or less manual handling, including pipetting, leading to variation in the final result. Also, manual interpretation of the analysis can result in diverse results.
Steps for reducing the variability of MS-based host cell protein identification
Two primary strategies are essential for addressing the complexity and variability of MS: Automation and normalization against internal standards.
- Automation: Pipetting robots are an excellent example of automating sample preparation to streamline multistep procedures and reduce variability from manual sample handling.
- Normalization against internal standards (proteins with properties similar to the target proteins): By spiking in proteins in known amounts and digesting them with the samples, we can use their results to correct enzyme activity and LC-MS performance variability. USP chapter 1132.1, currently in preparation, will contain information on suitable protein standards for MS analysis of host cell proteins.
At Alphalyse, the above approach has facilitated the monitoring of low-abundance Host Cell Proteins (HCPs) across diverse and complex drug types (recombinant proteins, mAbs, vaccines, Gene and Cell therapies), yielding coefficients of variation (CVs) below 20%.
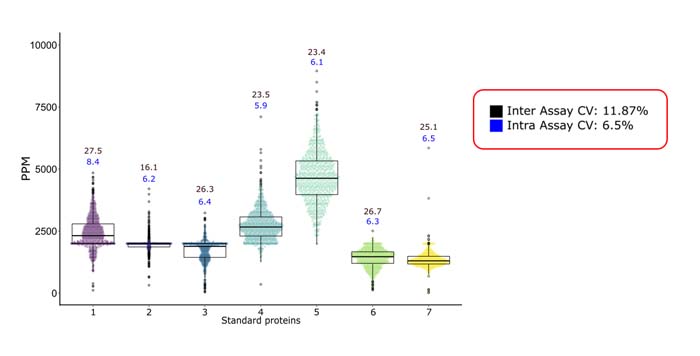
In conclusion, it is possible to achieve reproducibility in MS analysis by implementing automation and normalization strategies, facilitating its integration into QC laboratories, and ensuring reliable protein quantification.
Learn more in this webinar
If you are interested in learning more about how Alphalyses has achieved GMP certification for our MRM analysis of host cell proteins, you should watch this short webinar:

References
[1] Zhu-Shimoni et al.: “Host Cell Protein Testing by ELISAs and the Use of Orthogonal Methods, “Biotechnology and Bioengineering, 2014
[2] Bracewell et al.: “The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control, “Biotechnology and Bioengineering, 2015
[3] Wang et al.: “Host Cell Proteins in Biologics Development: Identification, Quantitation and Risk Assessment, “Biotechnology and Bioengineering, 2009
[4] Heissel et al.: “Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein,” Protein Expression and Purification, 2018
[5] The United States Pharmacopeia: “Residual Host Cell Protein Measurement in Biopharmaceuticals,” USP 39 – NF 34, 2016
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.
